

CONTENTS
Preface
Foreword (I): Sulfur metabolism - Yolande Surdin-Kerjan
Foreword (II): Sulfur nutrition and sulfur assimilation
of higher plants -
Christian Brunold
The plant sulfate transporter family: Specialized functions and integration
with whole plant nutrition -
Malcolm J. Hawkesford, Peter Buchner,Laura Hopkins and Jonathan R. Howarth
Molecular and metabolic regulation of sulfur assimilation: initial approach
by the post-genomics strategy -
Kazuki Saito
Metabolic regulation of cysteine synthesis and sulfur assimilation –
Rüdiger Hell
Regulation of sulfate uptake: from molecular bases to whole plant integration
- Lessons from research on the regulation of nitrate uptake -
Bruno Touraine, Timothy J. Tranbarger and Patricia Nazoa
Interactions between sulfur, nitrogen and carbon metabolism -
Christian Brunold, Peter Von Ballmoos, Holger Hesse, David Fell and Stanislav
Kopriva
S-Methylmethionine and the interface between sulfur and one-carbon metabolism
-
Sanja Roje and Andrew D. Hanson
Genetic and molecular analysis of phytochelatin biosynthesis, regulation and
function -
Christopher S. Cobbett
Physiological and environmental significance of selenium -
Danika L. Leduc and Norman Terry
Heavy metal regulation of cysteine biosynthesis and sulfur metabolism related
to stress in Arabidopsis thaliana trichomes - Cecilia Gotor, José R.
Domínguez-Solís, Gloria Gutiérrez-Alcalá, M. Carmen
López-Martín, Leticia Calo and Luis C. Romero
Roles of glutathione and glutathione-related enzymes in remediation of polluted
soils by transgenic poplars -
Tamás Kömives, Gabor Gullner and Heinz Rennenberg
Sulfur nutrition of deciduous trees at different environmental growth conditions
-
Cornelia Herschbach
Impact of volatile sulfur compounds on wine quality -
Doris Rauhut
Importance of sulfur for the quality of breadmaking wheat and malting barley
-
Fang-Jie Zhao and Steve P. McGrath
Contributed papers: (in alphabetical order)
Effect of N and S fertilization on the glucosinolate content of leaves and roots
of broccoli seedlings -
Alfredo Aires, Rosa Carvalho, Eduardo Rosa and Fernanda Maria Pereira
Screening of mutants accumulating high levels of anions and thiols from activation-tagged
lines of Arabidopsis thaliana - Motoko Awazuhara, Akiko Hayase, Masatomo
Kobayashi, Masaaki Noji, Mami Yamazaki, Motoaki Seki, Kazuo Shinozaki and Kazuki
Saito
Comparison of sulfur deficiency indicators in winter oilseed rape -
Ursula Balestra, Mechteld Blake-Kalff, J.-C. Lavanchy, A. Keiser, H.-R. Pfeifer
and D. Pellet
Further investigation on the role of the prolonged linker sequence in plant
sulfur transferases -
Michael Bauer and Jutta Papenbrock
Application of biacore technology for the analysis of the protein interactions
within the cysteine synthase complex -
Oliver Berkowitz, Markus Wirtz, Alexander Wolf, Jürgen Kuhlmann and Rüdiger
Hell
Influence of sulfur availability on non-protein thiols contents in transgenic
tobacco lines producing bacterial serine acetyltransferase -
Anna Blaszczyk and Agnieszka Sirko
Influence of the sulfur supply on the (iso)alliin content in leaves and bulbs
of Allium cepa and Allium sativum -
Elke Bloem, Silvia Haneklaus and Ewald Schnug
Model based prognosis of sulfur deficiency -
Elke Bloem, Silvia Haneklaus and Ewald Schnug
Glutathione transporter homologues from Arabidopsis thaliana -
Olivier Cagnac, Andree Bourbouloux and Serge Delrot
Sulfur status and acquisition in response to different sulfate concentrations
and time exposure in maize -
David Carden, Silvia Quaggiotti and Mario Malagoli
Nitrogen and sulfur requirement of Brassica oleracea L. cultivars -
Ana Castro, Ineke Stulen and Luit J. De Kok
Glutathione content and status of in vitro plantlets transferred to ex vitro:
demand for growth or response to oxidative stress? -
Luísa De Carvalho and Sara Amâncio
Sulfur dynamics in fallow soil and in the rhizosphere of field-grown rape and
barley -
Odile Dedourge, Phuy-Chhoy Vong, Françoise Lasserre-Joulin, Emile Benizri
and Armand Guckert
O-acetylserine (thiol)lyase confers tolerance to heavy metals -
R. Domínguez-Solís, M. Carmen López-Martín, M. Dolores
Ynsa, Francisco J. Ager, Luis C. Romero and Cecilia Gotor
Impact of atmospheric H2S on sulfur and nitrogen metabolism in Allium species
and cultivars -
Mark Durenkamp and Luit J. De Kok
Identification and characterization of Arabidopsis thaliana mutants resistant
to selenate -
Elie El Kassis, Nicole Cathala, Pierre Fourcroy, Pierre Berthomieu, Abderrahmane
Tagmount, Norman Terry and Jean-Claude
Davidian
The effect of catch crops on sulfate leaching and availability of sulfur in
the succeeding crop -
Jørgen Eriksen and Kristian Thorup-Kristensen
Characterization and differential expression of two sulfate transporters in
Arabidopsis thaliana -
Pierre Fourcroy, Elie El Kassis, Elisa Dalla Pria, Nicole Cathala and Jean-Claude
Davidian
Thiols in acorns and feeding mites collected at sites with naturally elevated
atmospheric sulfur concentrations -
Dieter Grill, Michael Tausz, B. Strnad, Astrid Wonisch, Maria Müller and
Antonio Raschi
Role of ATP sulfurylase in the regulation of the sulfate assimilation pathway
in plants -
Yves Hatzfeld, Nicole Cathala, Dao-Yao He and Jean-Claude Davidian
Investigation of the Fe-S cluster biosynthesis in higher plants -
Nicole Hausmann, Holger Eubel and Jutta Papenbrock
Transcriptome analysis of sulfur-starved Arabidopsis by DNA array: OAS is a
positive regulator of gene expression under sulfur deficiency -
Masami Yokota Hirai, Toru Fujiwara, Motoko Awazuhara, Tomoko Kimura, Masaaki
Noji and Kazuki Saito
Transcript profiling of sulfur depletion in Arabidopsis thaliana -
Rainer Höfgen, Victoria Nikiforova, Jens Freitag, Stefan Kempa, Kerstin
Riedel and Holger Hesse
Tissue and cell specific localisation of a sulfate transporter in maize -
Laura Hopkins and Malcolm J. Hawkesford
The role of elemental sulfur in tomato as a phytoalexin response to verticillium
infection -
Jonathan R. Howarth, Jane S. Williams, Richard M. Cooper and Malcolm J. Hawkesford
Selection of methionine-enriched soybean seeds -
John Imsande and M. Paul Scott
COR - an Arabidopsis thaliana protein with cystine lyase activity -
Patrik Jones and Kazuki Saito
New approaches to study "sulfur-induced resistance" against fungal
pathogens in Arabidopsis thaliana -
Ricarda Jost, Paul Scholze and Rüdiger Hell
Temporal and spatial expression analysis of serine acetyltransferase isoforms
in Arabidopsis thaliana -
Cintia G. Kawashima, Masaaki Noji and Kazuki Saito
The effect of lead on sulfoquinovosyl diacylglycerol content in leaves and roots
of wheat seedlings -
Oksana Kosyk, Alexander Okanenko and Nataliya Taran
Grass sulfur status and non-protein nitrogen accumulation: a preview in the
southeastern part of Belgium -
Richard Lambert, Michaël Mathot, Bernard Toussaint and Alain Peeters
Comparison of oilseed rape and barley rhizosphere microbial communities involved
in sulfur immobilization - preliminary results -
Françoise Lasserre-Joulin, Romain Boulan, Emile Benizri, Phuy-Chhoy Vong
and Armand Guckert
Identification and characterization of the AtNFS2 gene from Arabidopsis
thaliana encoding a nifs-like plastidial cysteine desulfurase -
SebastienLeon, Brigitte Touraine, Jean-François Briat and Stephane Lobreaux
Analysis of transgenic tobacco lines expressing bacterial CYSK gene encoding
O-acetylserine (thiol)lyase A -
Frantz Liszewska and Agnieszka Sirko
Effects of nitrogen availability on sulfate uptake in common reed -
Mario Malagoli, Claudia Bragato and Anna-Rita Trentin
Localization of g-glutamyl-transferase activity in plant tissue -
Antonio Masi, Tiziana Destro and Massimo Ferretti
Effects of nitrogen and sulfur fertilization on grass yield and quality in Belgium
-
M. Mathot, R. Lambert, J. Mertens, B. Toussaint and A. Peeters
Genomic and biochemical studies of sulfur assimilation in onion -
John McCallum, Meeghan Pither-Joyce, Martin Shaw, Anya Lambert and Michael T.
McManus
Effect of mineral sulfur and organic fertilizers on yield and sulfur uptake
of grass on Belgian sandy soils -
J. Mertens, G. Verlinden and M.Geypens
Arabidopsis sulfur transferases: investigation on their role in the organism
-
Tanja Meyer, Meike Burow, Michael Bauer and Jutta Papenbrock
Subcellular distribution of glutathione - a high resolution immunogold analysis
in leaves of pumpkin (Cucurbita pepo L.) -
Maria Müller, Bernd Zechmann, Michael Tausz and Günther Zellnig
Cluster analysis of gene responses to sulfur depletion in Arabidopsis thaliana-
Victoria Nikiforova, Carsten Daub, Jens Freitag, Stefan Kempa, Kerstin Riedel,
Holger Hesse and Rainer Höfgen
Altered expression of serine acetyltransferase gene in transgenic Arabidopsis
resulted in modulated production of cysteine and glutathione-
Masaaki Noji, Fumiko Saito, Tomoko Ochiai, Yumiko Shirano, Hiroaki Hayashi,
Daisuke Shibata, Tomohiko Kato, Satoshi Tabata and Kazuki Saito
Transgenic Arabidopsis thaliana expressing GFP in response to sulfur nutrition
-
Naoko Ohkama, Yoshitaka Sogawa, Derek B. Goto, Kentaro Takei, Hitoshi Sakakibara,
Nakako Shibagaki, Hiroaki Hayashi, Tadakatsu Yoneyama, Satoshi Naito and Toru
Fujiwara
Healthiness of winter oilseed rape fertilized with nitrogen and sulfur -
Anna Podlesna, Malgorzata Jedryczka and Elzbieta Lewartowska
Glutathione transport in pedunculate oak roots and its comparison with sulfate
and glutamine transport -
Heinz Rennenberg and Stefan Seegmüller
Successful engineering of methionine metabolism in potato -
Kerstin Riedel, Michaela Zeh, Anna Paola Casazza, Rainer Höfgen and Holger
Hesse
Effects of sulfur nutrition on carbon metabolism and nitrogen assimilation in
Chlorella sorokiniana -
Carmelo Rigano, Simona Carfagna, Vittoria Di Martino Rigano, Vincenza Vona,
Sergio Esposito and Graziella Massaro
Explaining sulfur and nitrogen interactions in the growth of cropped hybrid
ryegrass -
Maria Da Graça Serrão, Michael Tausz, Maria João Neves,
John Keith Syers and Fernando Pires
Selenate resistant mutants of Arabidopsis thaliana identifies a region
in sulfate transporter gene required for efficient transport of sulfate in roots
-
Nakako Shibagaki, Alan Rose, Toru Fujiwara, John P. Davies and Arthur R. Grossman
Sulfur uptake by rye grass as affected by different sulfur sources contained
in calcium nitrate -
Bal Ram Singh and Leif Ruud
Jungermanniidae species respond to cadmium in a different manner to
other bryophytes -
Kristin Sutter, Sieglinde Menge, Herbert Tintemann, Angelika Schierhorn and
Gerd-Joachim Krauss
Sulfur transport and assimilation in developing embryos of chickpea (Cicer
arietinum) -
Linda M. Tabe, Ingrid Venables, Anita Grootemaat and David Lewis
T-DNA insertion mutagenesis of sulfate transporters in Arabidopsis
–
Hideki Takahashi, Akiko Watanabe-Takahashi and Tomoyuki Yamaya
Modification of sulfur metabolism in spruce trees by H2S studied by radiolabelled
sulfate uptake -
Michael Tausz, Astrid Wonisch, Wilfried Weidner, Luit J. De Kok and Dieter Grill
Biosynthesis of cysteine and glutamate in Chlamydomonas reinhardtii:
effect of nitrate or sulfate starvation and cadmium stress -
José M. Vega, Antonio Benitez-Burraco, Javier Vigara and Carlos Vilchez
Mobilization of 35S in rhizosphere soil of rape and barley: relationship between
root-35S uptake and soil arylsulfatase activity -
Phuy-Chhoy Vong, Françoise Lasserre-Joulin and Armand Guckert
Wool quality and sulfur supply -
Shiping Wang, Yanfen Wang, Zuozhong Chen, Ewald Schnug and Silvia Haneklaus
Comparative biochemical characteri-zation of OAS-TL isoforms from Arabidopsis
thaliana -
Markus Wirtz and Rüdiger Hell
Effects of SO2 exposure on sulfur distribution in curly kale (Brassica oleracea
L.) Investigated by 35S-labelled nutrient solution -
Astrid Wonisch, Wilfried Weidner, Michael Tausz, Sue Westerman, Luit J. De Kok
and Dieter Grill
Interaction between atmospheric sulfur dioxide deposition and pedospheric sulfate
nutrition in chinese cabbage -
Liping Yang, Ineke Stulen and Luit J. De Kok
Characterization of two functional high-affinity sulfate transporters for uptake
of sulfate in Arabidopsis roots -
Naoko Yoshimoto, Hideki Takahashi, Frank W. Smith, Tomoyuki Yamaya and Kazuki
Saito
Influence of sulfur nutrition on sugar beet resistance to aphids -
Eva Zelená and Frantisek Zeleny
Influence of sulfur on growth of radish -
Frantisek Zeleny and Eva Zelená
Acknowledgements
Index of authors
Subject index
PREFACE
This book contains the invited and contributed papers of the 5th Workshop on
Sulfur Transport and Assimilation in Plants, a joined European Commission (COST
Action 829) and OECD meeting hosted at the Ecole Nationale Supérieure
Agronomique in
Montpellier (France) from April 11 to 14, 2002. The meeting was co-organized
by the ENSA-Montpellier (France), the University of Graz (Austria), the University
of Groningen (The Netherlands), Rothamsted Research, (United Kingdom), Institute
of Plant Nutrition and Soil Science, Braunschweig (Germany), the Agricultural
Biotechnical Center of Gödöllö (Hungary), Albert-Ludwigs-University
Freiburg (Germany) and the University of Chiba (Japan).
We are very pleased to dedicate this book to Prof. Dr. Christian Brunold, University
of Bern, Switzerland. His views and research have significantly contributed
to advanced understanding of the physiological, biochemical and molecular regulation
of the sulfur metabolism pathway and its interactions with nitrogen metabolism.
We also dedicate this book to Dr. Yolande Surdin-Kerjan, CNRS, Gif-sur-Yvette,
France, whose outstanding studies on genetics and molecular biology have contributed
to the identification of most of the genes involved in the regulation of the
sulfur metabolism pathway of the yeast Saccharomyces cerevisiae.
Jean-Claude Davidian
Dieter Grill
Luit J. De Kok
Ineke Stulen
Malcolm J. Hawkesford
Ewald Schnug
Heinz Rennenberg
editors
FOREWORD (I)
SULFUR METABOLISM
Yolande Surdin-Kerjan
Centre de Génétique Moléculaire, CNRS, 91198 Gif-sur-Yvette,
France
I entered the “Ecole Nationale Supérieure de Chimie de Paris”
with the intent of becoming a chemical engineer and then working in industry.
After graduation I was confronted with two facts: chemical industry was hostile
to the presence of women and anyhow, I was not good enough at it to consider
the life with chemistry. I thus went to visit Dr. Françoise Labeyrie
who was a friend of my family and whose laboratory was devoted to the study
of cytochromes. It took her only four hours to persuade me that Biology was
the research field to join. She introduced me to Dr. Huguette de Robichon-Szulmajster
who just created her research group to study the metabolism of amino acids deriving
from aspartate in Saccharomyces cerevisiae. Huguette wanted to use
S. cerevisiae as a model organism in a world then almost completely
devoted to E. coli. I was naive enough not to realize that, so this
point did not bother me, but I must say that, during my thesis work, I moderately
appreciated the remarks that I heard from time to time (“yeast ?, why
not rhinoceros…”). So, in September 1962, I got a position in CNRS,
and joined Huguette's laboratory.
It was only in 1969, after obtaining my Ph.D. that I switched to methionine
metabolism the study of which had been initiated by Hélène Chérest
(who still works with me). In 1974 Huguette died from a cancer without knowing
how good had been her intuition to use yeast as a model organism. I had to take
over and during the next 10 years, we characterized many genes involved in the
sulfur amino acids metabolism. More particularly we isolated the first yeast
strains impaired in sulfate transport. During these years also, I met Dominique
Thomas who came in the laboratory as a student and stayed. I thank him for staying,
for being so enthusiastic about the study of the regulation of transcription
of the sulfur amino acids metabolism, which he turned into a very sophisticated
model and for sharing my interest in metabolism.
In 1992, I met Jean-Claude Davidian. He phoned me to get our sulfate transporter
mutants, wanting to clone the plant sulfate transporter genes. Unfortunately,
I had lost them. He was disappointed but talked me into trying to isolate such
mutants again. He thus spent several months in my laboratory and, together with
Hélène Chérest, successfully characterized and cloned the
genes encoding the two yeast sulfate permeases. Meanwhile, Smith, Hawkesford,
Prosser and Clarkson published the isolation of a yeast mutant devoid of sulfate
transport. I still think they were paper, describing the isolation and characterization
of the two sulfate permeases from S. cerevisiae, was published after
theirs.
This is how I stepped into the world of plants. At the Montpellier meeting,
it was a great pleasure for me to realize that our yeast mutants have been successfully
used to clone several plant genes.
Sulfur amino acids metabolism in Saccharomyces cerevisiae
The biosynthetic pathway
Sulfur is an essential nutrient for all microorganisms. Extensive growth data
has been accumulated showing that S. cerevisiae possesses various enzymatic
systems that allow it to metabolize almost any sulfur source. In contrast to
many other microorganisms, yeast is able to use various organic sulfur compounds,
i.e. cysteine, methionine, homocysteine, glutathione, and S-adenosylmethionine
(AdoMet) as a sole sulfur source. This is due to the organization of the sulfur
pathway in yeast which allows the conversion of the main sulfur metabolites
into the others. This pathway has been almost completely deciphered by my research
group, using first biochemistry and classic genetics and later, the powerful
reverse genetics methods. Finally, our work has also been greatly helped by
the completion of the sequence determination of the yeast genome in 1996. To
date, we have characterized more than 15 genes encoding enzymes of the sulfur
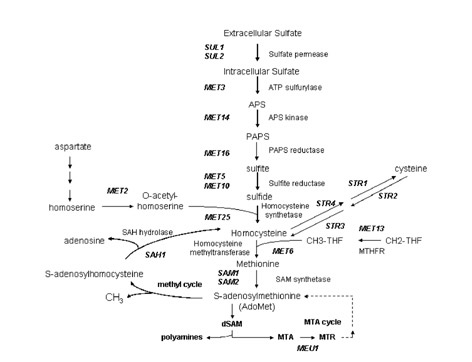
amino acids pathway (Fig. 1).
In addition, we have shown that some mutants requiring methionine for growth
bear mutations in genes encoding enzymes not directly related to the sulfur
amino acids metabolism. These genes are MET19, MET22 and SOD1,
encoding glucose-6- phosphate dehydrogenase, 3',(2'),5'-bisphosphate nucleotidase
and one superoxide dismutase, respectively, as well as the less well characterized
MET18 and MET27 genes. It is still unclear why mutations within
these genes result in methionine auxotrophy, but it suggests that some functions
of sulfur amino acids remain to be uncovered (Thomas and Surdin-Kerjan 1997).
Recently, we have also characterized the FOL3 and the MET7
genes encoding the dihydrofolate synthetase and the folylpolyglutamate synthetase
respectively, both involved in the addition of glutamate tails to folate coenzymes
(Cherest et al. 2000). The fol3 and met7 mutants have been
used recently to characterize the corresponding plant genes (Ravanel et al.
2001).
Recycling of the products of the catabolism of AdoMet
Methionine is not only involved in protein synthesis but is also an essential
determinant of the one carbon metabolism. Indeed, under its activated form,
Sadenosylmethionine (AdoMet), it is the methyl donor in numerous transmethylation
reactions of nucleic acids, proteins or lipids. Further, AdoMet serves as a
precursor for the biosynthesis of polyamines and is one of the substrates used
in a number of reactions, including vitamin biosyntheses and nucleotide modifications.
 ^
to the top
^
to the top
Fig. 1. Metabolism
of sulfur amino-acids in Saccharomyces cerevisiae. APS, adenosine 5'-
phosphosulfate; PAPS, adenosine 3'-phosphate 5'-phosphosulfate; MTHFR, methylene
tetrahydrofolate
reductase; CH3THF, methyl-tetrahydrofolate; CH2THF, methylene tetrahydrofolate;
MTA,
methylthioadenosine; MTR, methylthioribose-1-phosphate; dSAM, decarboxylated
AdoMet.
Given such ubiquitous functions, the equilibrium between methionine and AdoMet
is thus expected to be of crucial importance for the overall cellular homeostasis.
In eucaryotic cells, the methionine/AdoMet ratio was thought to be largely controlled
through two recycling pathways that act on the products of AdoMet catabolism.
The first one, called the methyl cycle, allows the conversion of Sadenosylhomocysteine,
the by-product of all transmethylation reactions, into homocysteine which is
next remethylated into methionine by the methionine synthase. The second one
comprises a set of complex reactions that allow the direct synthesis of methionine
from 5'-methylthioadenosine (MTA), a compound formed mainly during polyamine
biosynthesis. In this pathway (called the MTA cycle), the ribose moiety of the
adenosyl group gives rise to the four carbon skeleton of methionine while conserving
the methylthiol group (Fig. 1).
Using molecular genetics, we have been able to prove that the methyl cycle as
well as the MTA cycle are both active in S. cerevisiae. In addition, more recently,
we have uncovered a pathway allowing the utilisation of S-methylmethionine by
yeast, and we have shown that AdoMet can be used to directly methylate homocysteine,
yielding methionine in what seems to be a futile cycle. However, this cycle
is fully active when AdoMet is the sole sulfur source present in the growth
medium (Thomas et al. 2000).
Transport of sulfur compounds
Transport systems allow yeast cells to extract virtually any sulfur compounds
from their environment. Using specifically designed genetic screens, we have
isolated and characterized: (i) the genes encoding the two sulfate transporters,
Sul1p and Sul2p (Cherest et al. 1997). As I mentioned, the SUL1 gene
was isolated first by Smith et al. (1995); (ii) the two methionine transporters,
Mup1p and Mup3p (Isnard et al. 1996); (iii) the AdoMet transporter, Sam3p (Rouillon
et al. 1999); and (iv) the Smethylmethionine transporter, Mmp1p (Rouillon et
al. 1999).
In addition to these highly specific transport systems, biochemical uptake assays
have revealed the presence in yeast of less specific transport systems for most
of the sulfur compounds.
Regulation of sulfur amino-acids biosynthesis
The first observations about the MET regulatory system were made in
the 1970's. We showed that when wild type S. cerevisiae cells are grown
in the presence of a high concentration of methionine (1 mM), a decrease of
the synthesis of all the enzymes implicated in methionine biosynthesis is measured.
Later, we were able to show that this negative regulation is acting at the transcription
level and that the signal was AdoMet. Then, we identified several cis-acting
regulatory sequences found upstream of the MET genes and demonstrated that the
negative regulation resulted from the non-activation of the transcription of
the genes (Thomas et al. 1989).
During the 1990s, we isolated most of the genes encoding the factors responsible
for the transcriptional regulation the MET genes, i.e. Met4p, Met28p,
Met30p, Met31p and Met32p. Meanwhile, Cbf1p, a protein that functions at both
centromeres and MET gene promoters was characterized in several laboratories
and its precise role in MET gene regulation was deciphered in my laboratory.
All these factors were identified either by specific genetic screens or through
specific molecular assays, such as one hybrid experiments.
The identification of so many factors was suggestive of an unanticipated complexity
of the molecular mechanisms underlying the regulation of the MET gene
network. We have been able to decipher the precise function of all these factors.
Briefly, a unique transcriptional activator, Met4p, is recruited to the MET
promoters trough the assembly of different large multiprotein complexes (Cbf1p-Met28pMet4p
and Met4p-Met28p-Met31p (Met32p)) (Kuras et al. 1996; Kuras et al. 1997; Blaiseau
and Thomas 1998) Indeed, the binding of Met4p to DNA varies from one gene to
another. Once tethered to DNA, Met4p activates the transcription of the downstream
gene owing to an unique acidic activation domain. When the cells are exposed
to a high methionine concentration, Met4p is inactivated through the activity
of the SCFMet30 ubiquitin ligase and MET gene transcription is turned
off. SCF ubiquitin ligases are multi-protein complexes, that use substrate-specific
adapter subunits termed F-box proteins (such as Met30p) to recruit substrates
for ubiquitylation by a core apparatus, which is composed of the scaffold protein
Cdc53/cullin, the RING finger protein Rbx1, the adapter protein Skp1, and the
E2 enzyme Cdc34. Surprisingly, the consequence of Met4 ubiquitylation differs
according to the growth conditions. In cells grown in minimal medium, Met4 ubiquitylation
triggers its degradation by the 26S proteasome (Rouillon et al. 2000). In contrast,
in rich medium, ubiquitylated Met4p is stable but unable to bind to the MET
promoters while ubiquitylated Met4p is still capable of activating the AdoMet
biosynthesis genes. Thus, ubiquitylation not only regulates the MET
gene network by distinct degradation-dependent and -independent mechanisms,
but also controls the differential recruitment of Met4p, thereby diversifying
its activation specificity (Kuras et al. 2002).
I am very grateful to the editors and more particularly to Jean-Claude Davidian
for partly dedicating this volume to me. And I am happy to share this honour
with Dr. Brunold whom I have learned to know during these last three sulfur
meetings and whose great knowledge of plant sulfur metabolism I have come to
appreciate. This honour is a great reward and well compensates me for the hard
work during all these years. I think that I have lived during a fantastic period
of time during which, starting with a partially known biosynthetic pathway,
we could end by formulating a recognized model for the regulation of transcription.
This was greatly helped by our knowledge of the metabolism itself. With the
modern techniques that can be now used for plant metabolism, I hope that the
young scientists will live rewarding moments as I did.
Acknowledgements
I thank my friend Professor Pete Magee for editing this manuscript and for being
always present to help me when needed, ever since we met in Huguette's lab in
1965.
FOREWORD (II)
SULFUR NUTRITION AND SULFUR ASSIMILATION OF HIGHER PLANTS
Christian Brunold
IPS, University of Berne, Altenbergrain 21, 3013 Bern, Switzerland
H2S as a starting point
I wish to thank the organizers of the 5th International Workshop on Sulfur Nutrition
and Sulfur Assimilation in Higher Plants for their kind dedication of this distinguished
volume containing many important contributions to the advancement of our knowledge
of sulfur metabolism to Yolande Surdin-Kerjan and myself.
Sulfur metabolism was the main field of my scientific work since I started my
Ph.D. thesis in 1969. At that time, my adviser, Prof. K.H. Erismann, suggested
to find out, if higher plants could use electrons from H2S instead of H2O for
producing reducing equivalents in photosynthesis. Since I was more interested
in metabolism than in electron transport, I proposed to use H2S as sulfur source
and to analyze, if it had a regulatory effect on sulfate assimilation. At that
time we knew that Spirodela oligorrhiza (Ferguson 1969), but also Lemna
minor, the organism preferentially analyzed in Erismann's laboratory, used
NH4 + rather than NO3 - as a nitrogen source, when both ions were available
in the nutrient solution. With this knowledge in mind, the obvious working hypothesis
was that H2S rather than SO4 2- would be used as a sulfur source, when both
were available. L. minor turned out to be cooperative, did exactly
what we expected, and thus made it possible to finish the thesis in 1972. The
results obtained can be summarized as follows: In L. minor gassed with
6 ppm H2S, uptake and assimilation of sulfate were almost completely inhibited.
H2S taken up was partly directly used for cysteine formation, thus increasing
the cysteine content (Brunold and Erismann 1975) and partly oxidized, which
increased the sulfate content. The work with H2S treated L. minor had
several very positive consequences, since (i) I was offered a position at the
Institute of Plant Physiology at the University of Berne, Switzerland, and could
start an academic career, (ii) I got a grant from the Swiss National Science
Foundation (SNF) for working in the laboratory of Jerry Schiff at Brandeis University,
which led to an inseparable tie with the field of sulfur metabolism, and to
my first publication in an international plant journal, (Brunold and Schiff
1976), (iii) I got into contact with Ineke Stulen and Luit de was contacted
by Ahlert Schmidt for a successful collaboration (Brunold and Schmidt 1976),
and (v) I met Dr. Bergmann, University of Cologne, Germany, who brought me into
contact with Heinz Rennenberg, with whom I collaborated till the end of my scientific
career (Rennenberg and Brunold 1994; Hesse et al. 2003). Looking back, I consider
it a great privilege that Dr. Erismann allowed me to work in sulfur metabolism
for my Ph.D. thesis.
Answers and questions
After studying the regulation and the localization of sulfate assimilation for
many years, I now make the experience many scientists made before me: There
are more open questions, than when I started. This situation can be well exemplified
using adenosine 5'-phosphosulfate (APS) reductase (APR, Setya et al. 1996; Suter
et al. 2000), formerly named APS sulfotransferase (Schmidt 1972), my favored
enzyme:
1. Like sulfate transporters (Takahashi et al. 2000; Vidmar et al. 2000), APR
is especially sensitive to a lack (Brunold et al. 2002) or a surplus of reduced
sulfur (Brunold and Schmidt 1976; Westerman et al. 2001; Vauclare et al. 2002).
There are several indications (Takahashi et al. 2000; Vidmar et al. 2000; Vauclare
et al. 2002) that GSH plays a role in signaling the sulfur status of plants,
however, the genetic sequences, the genes and the mechanisms involved, which
lead to induction or repression of APR during lack and surplus of sulfur, respectively,
are not known.
2. The assimilation of sulfate and nitrate is regulated reciprocally in a coordinate
manner (Brunold et al. 2002). O-acetyl-L-serine (OAS) seems to play a role in
this coordination (Neuenschwander et al. 1991; Harms et al. 2000), but the mechanisms
involved are not known. This is in contrast to the situation in bacteria, where
regulatory mechanisms involving N-acetyl-L-serine (NAS), an isomer of OAS, have
been elucidated in great detail (Kredich 2000), which can give indications for
analyzing plant systems. APR may be the enzyme of choice for this analysis,
because it is very sensitive to regulatory signals and is easy and fast to measure.
3. APR is regulated by sugars (Kopriva et al. 1999) and several mechanisms,
which might be involved, have been described (Hesse et al. 2003). Regulation
by sugars is also important in other systems, therefore, checking proposed mechanisms
(Smeekens 2000; Hesse et al. 2003) and possibly new ideas using APR as an indicator
of the sugar status may result in contributions of general relevance in the
field of plant sugar sensing.
4. Calculation of the flux control coefficient of APR in A. thaliana
root cultures indicated that APR had a high flux control coefficient, but was
not rate limiting (Vauclare et al. 2002). The question is, if corresponding
calculations in other systems, especially in green tissues, result in similar
values for APR and in addition give values for the flux control coefficients
of other enzymes of sulfate assimilation. Such a mathematical analysis would
greatly help to understand the enzymatic regulation of this pathway.
5. In maize leaves, APR is localized exclusively in bundle sheath cells, and
cysteine formed there is transported into the mesophyll cells, the predominant
cell type for GSH synthesis (Burgener et al. 1998). Several ideas for explaining
this special localization have been discussed, but an unequivocal physiological
explanation is still missing. The question is complicated by the fact that the
localization of sulfate assimilation in bundle sheath cells as detected in some
C4 grasses is neither a prerequisite nor a consequence of C4 photosynthesis,
since in dicot C4 Flaveria species, APR mRNA and protein were present
at comparable levels in both types of cells (Koprivova et al. 2001).
6. It seems clear now that higher plants use APS as a sulfonyl donor in sulfate
reduction, bacteria either use APS or adenosine 3'-phosphate 5'-phosphosulfate
(PAPS), and in the moss Physcomitrella patens both APS and PAPS dependent
sulfate assimilations coexist (Koprivova et al. 2002). The physiological, ecological
and evolutionary causes for these different choices of sulfonyl donors are not
known.
Scientists, Problems and Money
The interests of the scientists working in a certain field are changing with
time, leaving some questions unanswered, focusing on others and possibly, but
not necessarily, coming back to the old ones after some time for testing new
ideas and new methods. It is therefore possible that at least some questions
of the small selection presented above will never be answered, but answers to
some of them will certainly be presented soon, because they are not only intriguing
for me, but for many colleagues, too. At the same time I look forward to answers
to interesting problems from other colleagues of the “sulfur family”.
I use this term, because over the years, the scientists working on sulfur metabolism
of higher plants have developed into a community, in which the individuals do
not only know and appreciate each other scientifically, but also personally.
From my point of view, this development is mostly due to Ineke Stulen and Luit
De Kok, who first initiated the Sulfur Workshops, and later on started COST
action 829. This is my last occasion for thanking them for all what they did
to promote research on sulfur metabolism. I consider it a great privilege that
I got acquainted with both of them long ago, that I could learn a lot from them
and that I could feel their sympathy and friendship.
At the beginning of my work on sulfur metabolism, I was not interested in the
practical application of my findings, but in basic research. I wanted to know
how sulfur assimilation was regulated and where it was localized. In my grant
applications, of course, I always stressed that research on plant metabolism
in general and on sulfate assimilation in particular was very important, because
plants are the absolute basis for nourishing men, and sulfur metabolism was
not only essential for the synthesis of storage proteins, but also for coping
with many stress situations encountered by plants (Rennenberg and Brunold 1994).
My grant applications were accepted, I do not know, however, if the granting
committees were especially impressed by my general ideas about the importance
of plant metabolism.
Today, more than 800,000,000 people are suffering from hunger, and the first
symptoms of climate change with its stressful consequences for plants become
evident. Therefore basic knowledge about plant metabolism is more important
than ever, but plant scientists have now the additional obligation to contribute
to solving practical problems. In this respect, COST actions and EU projects
in general represent ideal platforms, and this will even be more the case, when
COST is transformed into a system, in which all participating groups are subsidized
with grant money. Up to now, granting COST actions was a Swiss specialty. This
made the task of the Swiss coordinator easy to bring excellent Swiss groups
together for working in a COST action and the future funding will also make
the job of the European coordinator a more pleasant one. At the present time,
I am happy that 5 Swiss research groups, funded by the Swiss Agency for Education
and Science, are working in the “sulfur” COST 829 and that two Swiss
groups are participating in a “sulfur” EU project. I am especially
happy about the Swiss participation in this EU project, because my successor,
Doris Rentsch, joined this excellent consortium of groups.
I would like to thank my friends Luit De Kok, Dieter Grill, Malcolm Hawkesford,
Heinz Rennenberg, Kazuki Saito, Ewald Schnug, Ineke Stulen and Ervin Balazs
for organizing this workshop. My special thanks go to Jean-Claude Davidian,
because his friendship and his kind personality gave this event a touch of warm
humanity.